Strain Requirements
The following checklist can be used as a quick reference to ensure your strains meet our submission criteria.
Samples that do not meet one or more of the criteria below can be refused, or fail to sequence entirely, so please ensure you read the entire document to avoid any issues or delays.
Tubes: |
Receive tubes from us |
Sample ID Labels: |
Affix sample ID labels lengthwise |
Sample Info: |
Upload to project portal |
Culture: |
Grow your bacteria |
Transfer: |
4 x 106 - 6 x 109 cells or 30 - 50 mg |
Ship: |
Package and label correctly |
We will NOT be able to accept your samples if you have not entered the sample information in your project portal.
The full protocol available in PDF format has additional information, including a visual summary, a worked example on calculating optical density, and some tips and tricks to optical density measurements.
Overview
Preparing bacteria for submission to MicrobesNG is easy - we provide tubes containing a buffer, which inactivates the cells and stabilises the DNA. The buffer also effectively inactivates pathogens so you can send bacterial samples from around the world, hassle free. GMO’s and CL/BSL-3 strains accepted Ship at ambient temperature, through regular post
We do not grow your bacteria - instead, we extract DNA directly from the material that you send. For this reason, you need to send between 4 x 109 to 6 x 109 cells, or a pellet that weighs between 30 and 50 mg when wet.
This protocol will give you some pointers on how to:
- Grow a pure culture
- Quantify the number of cells
- Harvest the cells
- Ensure that the ratio of cells to buffer is correct so that all the cells are properly inactivated, and the DNA is stabilised
Inactivation Buffer
The tubes that we send you contain 500 µL of DNA/RNA Shield from Zymo Research. This buffer effectively inactivates pathogens, including tough-to-lyse microbes, without the need for any additional steps. Samples are stable at 4-25˚C for >2 years, and indefinitely at -20˚C. When exposed to low temperatures, the buffer may crystalise. This can be resolved by warming the buffer to room temperature and mixing.
We ask you to read the Safety Data Sheet for DNA/RNA Shield, which can be accessed via the manufacturer’s website.
If you prefer to provide your own tubes, we require 2 mL screw cap tubes with an outer diameter of 11 mm, which is the standard diameter for microcentrifuge rotors. Tubes must contain 500 µL of DNA/RNA Shield, which can be purchased from Zymo, or one of their regional distributors. You then can print your sample ID labels from your project portal; note that we do not accept handwritten labels.
Sample Information
Before sending your samples, it’s vital that you submit your sample information in your online project portal. We need to know the species of the organisms you are sending to ensure we can meet our guarantees on coverage, and provide the best QC possible.
If you do not know the species, we can accept identification to genus level, however this may impact the QC of your sequencing data. If you don’t know the identity of your organism, then please contact us at info@microbesng.com.
We will not be able to accept your samples if you have not entered the sample information in your project portal.
Growing your cells
We all know how to grow bacteria, but here are some steps to ensure you send a pure, clonal bacterial strain. Purity is important for obtaining the highest quality assembly.
1. Prepare an agar plate of the strain you wish to sequence.
2. Visually inspect the plate to ensure there is a single phenotype.
3. Pick a single colony and subculture either in liquid broth, or on an agar plate.
If you grow your cells in liquid broth, we recommend that you resuspend a single colony in 200 µL of a suitable sterile buffer (e.g. PBS), and use 100 μL as an inoculum. Following culture, we would recommend plating an aliquot of this fresh growth to ensure no contamination occurred during the process.
If you grow your cells on agar plates, aim to have a lawn covering between ½ to ⅔ of the plate, and streak the rest of the plate to obtain single colonies to check the culture is pure. Depending on the species you may need to prepare more than one plate.
Quantifying your cells
There are several ways of quantifying bacterial growth. This protocol uses Optical Density at 600 nm (OD600), which is suitable for many organisms - but not all. This method requires access to a spectrophotometer, and we recommend that you follow the relevant instruction manual for your specific instrument to ensure you obtain accurate readings.
If OD is not a good fit for your organism, you can also centrifuge your bacteria and send us a pellet that weighs between 30 and 50 mg when wet. Alternatively, you might consider plate counts of serial dilutions (e.g. Miles & Misra) or flow cytometry.
If you subcultured your pure single colony into a liquid broth, you should aim to harvest cells during the mid log/exponential growth phase. This is typically between an OD600 of 0.5 - 1, depending on the organism you are culturing. Mix the culture thoroughly before measuring the optical density and blank using sterile broth.
If you grew your bacteria on an agar plate, create a bacterial suspension by harvesting ½ to ⅔ of total fresh growth from the plate with a sterile loop and resuspend it in 5 - 10 mL of a suitable sterile buffer (e.g. PBS). You can then blank the spectrophotometer with an aliquot of the sterile buffer, and measure the optical density of an aliquot of your bacterial suspension.
OD measures are only valid when they are below 1.0, so you may need to prepare a serial dilution of your bacterial suspension, with the series continuing until one of the dilutions has an OD below 1.0. You can then calculate the OD of the stock culture by multiplying the measured value by the dilution factor.
Harvesting cells
We require 4 - 6 x 109 cells - this ensures that the cells are inactivated and safe for transport and handling. A quick way to determine the number of cells is to use an equivalent OD of 8 - 12.
For example, if you’re harvesting a liquid culture at an OD600 of 0.8, you would need to process 10 - 15 mL. If the stock bacterial suspension you prepared from an agar plate has an OD600 of 80, you would need 0.10 - 0.15 mL.
Take an appropriate volume and centrifuge to pellet the bacterial cells. Discard the supernatant, and wash the pellet with 1 mL of sterile buffer (e.g. PBS), before centrifuging a second time.
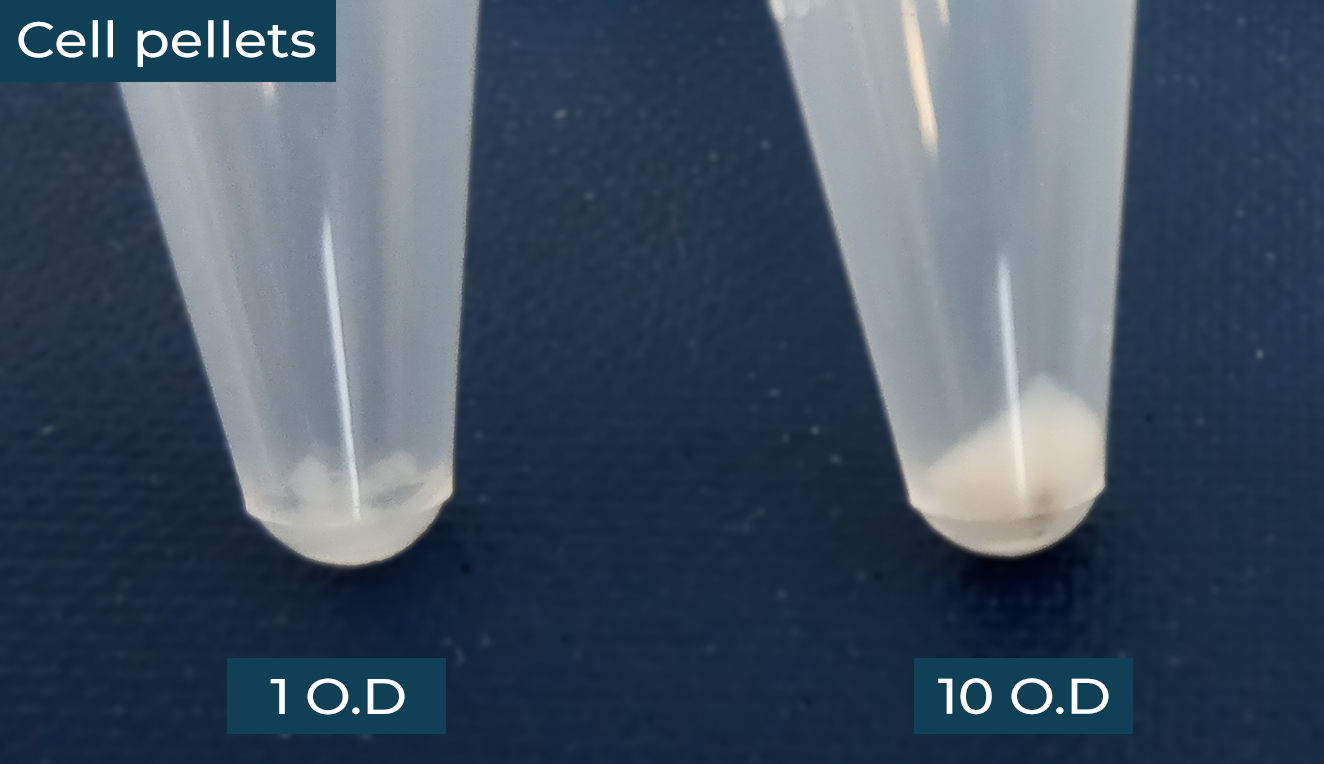
Figure 1. Pelleted S. aureus cells; the right tube contains cells at an equivalent OD600 of 10, or ~40 mg in wet weight. The pellet on the left shows an example of equivalent OD600 of 1, which is insufficient for submission.
Take 0.5 mL of the inactivation buffer from the sample tubes provided to you, and resuspend the bacterial pellet.
Transfer this resuspension back into your sample tube, ensure the lid is tightly fastened, and affix the MicrobesNG Sample ID label we sent with your tubes.
Please attach labels to your sample tubes lengthwise, not around the tube (Figure 2).
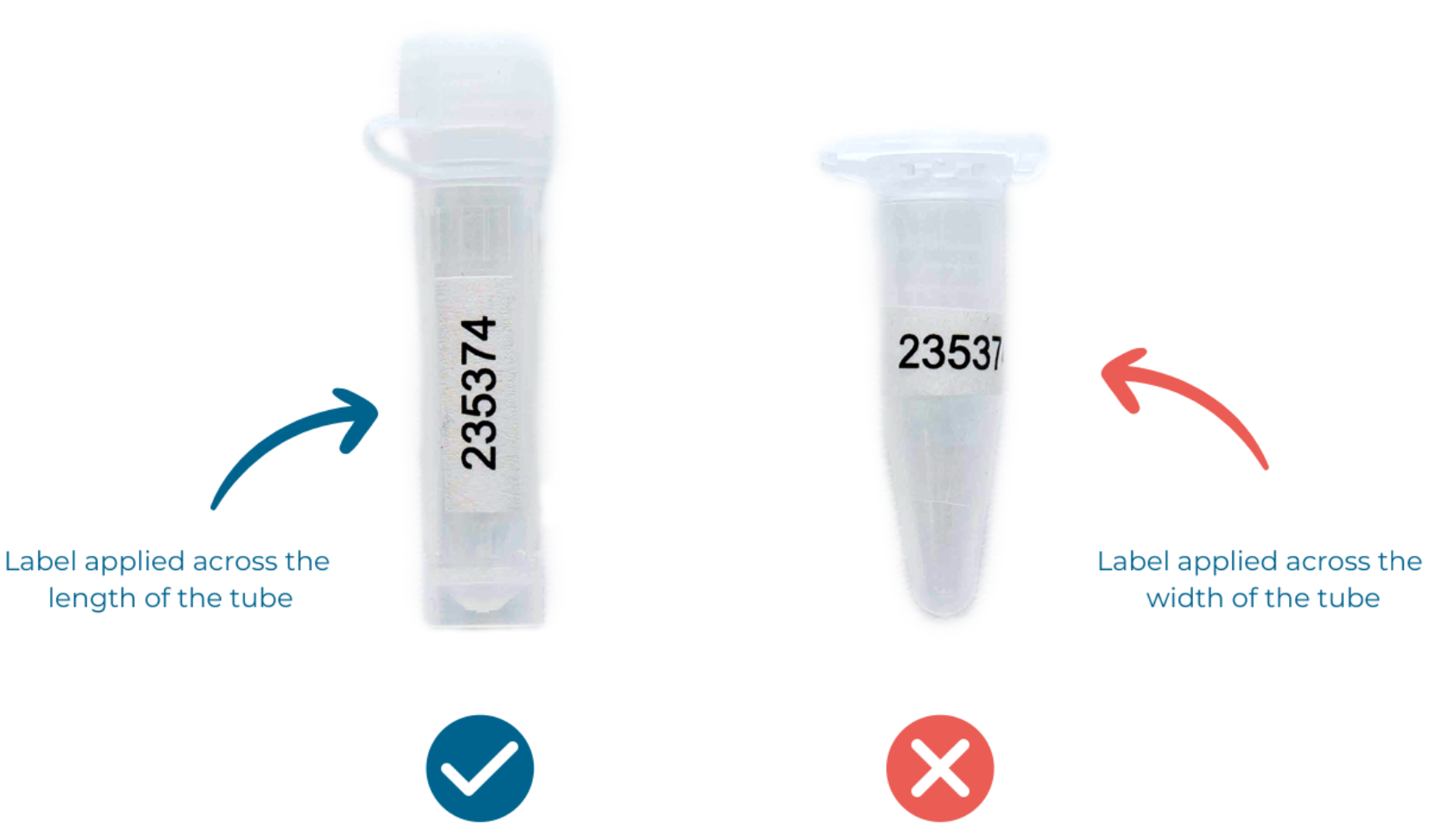
Figure 2. How to correctly apply labels to your tubes. Please use the tubes we provide, or if supplying your own, ensure they are 2 mL thread capped tubes containing 500 µL of DNA/RNA Shield Inactivation Buffer.
We will not be able to accept your samples if you use incorrect tubes or labelling, or if you have not entered the sample information in your project portal.
Shipping
If you expect your shipment to arrive within 5-7 days, the samples can be sent at ambient temperature. If your package will be in transit for longer than this, we recommend including ice packs.
To prevent your samples from leaking during shipment, place the tubes in a rigid tube box, with individual spacers. Placing your tubes in the box in order of the sample ID number greatly reduces handling, and will give you the best possible turnaround time.
If we sent your buffer tubes in a plastic box, please use this to ship your samples to us.
If you are sending more than one project in the same shipment, please use a separate box for each project and clearly label each box with the project number. If you are not able to use a box, please separate tubes from different projects in another way, e.g. inside a falcon tube or zip lock bag.
Place the rigid tube box inside an outer package (e.g. polystyrene or cardboard box) along with an absorbent material to catch any leaks, and protect it with suitable packaging material.
Log onto your project portal to print the return label and attach it to the outside of your package. Using this label will help our Laboratory Team to identify your project and ensure your samples enter the correct pipeline as quickly as possible. We recommend that you send your parcel using a tracked service.
Make sure that you have entered the sample information in your project portal before shipping your samples! We use this information to optimise the extraction process.
We will NOT be able to accept your samples if you have not entered the sample information in your project portal.