Top 5 most commonly asked questions
- I haven’t received an email / quote
We have a very good response time for enquiries, so if you haven’t heard back from us you may be experiencing an issue with receiving our emails. Please check your Junk and Quarantine folders.
If you cannot receive our emails, we ask that you contact your IT department and request that they add the following domains to their allow list:
- Microbesng.com
- Plasmidsng.com
- Xero.com
- How can I send my samples to you?
The most important part of getting your samples to us is ensuring you've followed our sample preparation protocols correctly.
You'll find details on how to send your samples to us in the protocol relevant to your service.
- What is your turn-around time?
We calculate turnaround time from the point we receive your samples and accept them into our sequencing queue.
Our target turn-around time is 5-7 weeks for our Short, Hybrid, Scout and Pioneer (metagenomics) services.
Our Long Read service has a target turnaround time of 4 weeks.
If you have a specific requirement for project completion, get in touch with us at info@microbesng.com.
- Can I get a copy of your sample submission protocols?
You can find all of our submission protocols at microbesng.com/documents
NOTE: Please do not send us any samples before you have received your sample ID numbers
It’s really important that you follow our preparation protocols to ensure the safety of our laboratory staff, and the integrity and provenance of your samples. Failure to comply with our requirements may result in your samples being destroyed.
If you have any questions, drop us a line at info@microbesng.com and include your project reference number if you have one.
Quotes, Ordering and Payment
- How do I get a quote and pay for my order?
Please request a quote online, and put anything we need to be aware about in the comments section.
We’ll send your quote out within 2 working days. If you have not heard from us in that time, please check your junk mail and quarantine folders, and if you still can’t find any correspondance, contact us at info@microbeng.com.
If you want to proceed with your order, there are currently two ways to pay:
Online Payment
You can pay online via credit card. After you accept your quote, we’ll send you a link along with your invoice. We accept all major credit and debit cards.
Invoice
If you would like to receive an invoice in relation to your order please forward us a Purchase Order document (with full contact information of your Accounts Payable Department and VAT registration number) to info@microbesng.com.
If you would like to receive an invoice, but your institution does not use a PO system we require the following information:
- Full institution name, postal address and email address for where you would like the invoice sent.
- VAT registration number.
- Any grant code / reference number that you need to appear on the invoice.
Please note that MicrobesNG is our Trading name and Microbial Genomics Ltd is our Registered name. Your purchasing system may have us set up under either of these names.
You can only send your samples when we have received payment and you have been given your unique sample ID numbers.
- Do you offer discounts?
Yes! We have excellent volume discounts available.
If your colleagues work with microbial genomes you can always pool money together to save a bit of cash!
Please see our Whole Genome Sequencing service page for pricing.
- Can you handle high volume orders of over 1000 samples?
Yes! We have a proven track record of delivering high volume orders on time, providing high quality data and an excellent customer experience.
Working with extremely large volumes? drop us a line on info@microbesng.com and our dedicated business development and key accounts team will contact you.
- Why register as a user on your website?
You can register as a user by clicking at the top right tab on our website “Register”.
By doing this, you will be able to see all of the different projects you have with us, the status of quotes, and if you are the primary contact of a project, you will also be able to add/ remove collaborators and grant them access to that project.
Please make sure to use the exact same email address that you have used to communicate with us and to request quotes.
If you log on before requesting a quote it will also autofill your details, saving you a bit of time!
Whole Genome Sequencing
- What sequencing platforms do you use?
We sequence our Illumina based services on a NovaSeq 6000 using 2 x 250 bp kits, and our Oxford Nanopore Technologies based services on a GridION with R10.4.1 flowcells.
Find out more in our methodology document!
- What’s your policy for resequencing failed samples?
You can find details on our resequencing policy here. In summary, as long as your sample meets our QC thresholds, and is free from contamination, we will resequence your sample one additional time at no charge.
We may ask for you to resubmit samples if the original sample did not meet our QC criteria, or if we have insufficient volume for an additional library preparation.
- What’s the difference between Short Read, Long Read and Hybrid? Which should I use?
We have a short video that will help explain, which you can watch here.
Our short read service sequences 250 base pair fragments of your genome. These fragments are overlapped and joined together in an assembly, but there are limits to how well a genome can be reconstructed from short fragments alone.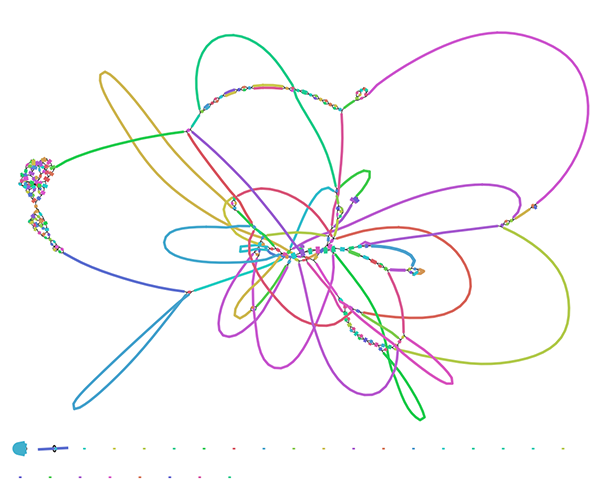
This is a typical short read assembly graph. But what happens if you use long reads?
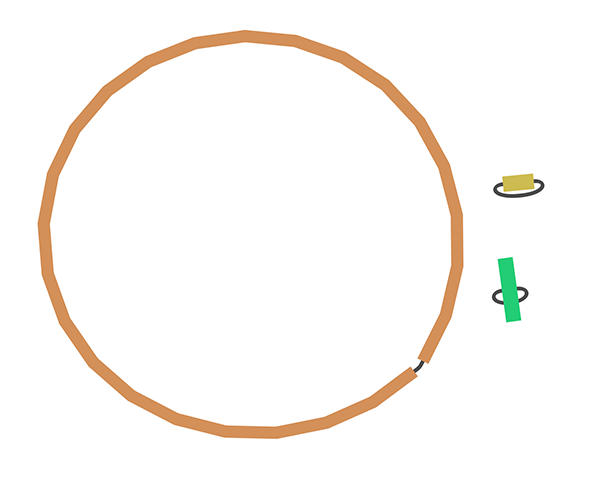
That's better!
That's because we start with much longer fragments that are easier to piece back together, allowing reconstruction of whole chromosomes and plasmids.
But short reads are still useful; they’re a lot more accurate, so our Hybrid service combines both Short and Long reads to get the best of both worlds, generating reference quality assemblies.
- What depth of coverage is provided? Can I get additional coverage?
Our Short Read service has a target coverage of 30X, and our Long Read service has a target coverage of 50X. Hybrid offers 30X Short Reads, and 50X Long Reads.
You can expand the coverage of our Short Read service for deep sequencing and population sequencing applications. We charge £32.50 Ex. VAT for additional 30X coverage units, which can be expanded all the way up to 510X. This can be configured on our quote form, where you will see live pricing.
We don’t currently offer expanded coverage for our Long Read or Hybrid services on our quote form, but if this is something you are interested in drop us a line at info@microbesng.com.
- Do you offer amplicon sequencing, RNA-seq, ChIP-seq, TraDIS-seq, human, animal, extra-terrestrial or other non-microbial sequencing?
MicrobesNG’s focus is on being the best place to do your microbial whole genome sequencing and metagenomics projects, and we are not able to process other sample or experiment types.
- Do you offer plasmid sequencing?
Yes! You can access PlasmidSeq from our sister company PlasmidsNG. PlasmidSeq is a rapid turnaround service that provides the full sequence of your plasmid for just £10 Ex. VAT.
Metagenomics Sequencing
- What’s the difference between 16S and Shotgun Sequencing?
Sequencing strategies like 16S rely on specific amplicons to identify the constituent taxa present in a DNA prep. This means you are limited to information pertaining to that amplicon only, and are beholden to some of the quirks that can entail.
We prefer full fat metagenomics, in the form of shotgun sequencing, which captures all of the DNA in your sample. This lets you identify a wider range of organisms, and get additional genetic information that 16S would miss.
- What are the Metagenomics services offered by MicrobesNG?
Whilst 16S has its limitations, it is cost effective. We wanted to offer a higher resolution of analysis than 16S while keeping cost efficacy in mind. That’s where our metagenomics services Scout and Pioneer come in.
- Scout - minimum 1 million reads (at least 0.5 Gb)
- Pioneer - minimum 2 million reads (at least 1.0 Gb)
We want the services to be useful to experienced metagenomics customers who want to see the diversity/quality of their sample before embarking on deeper sequencing.
We also want the services to be accessible to customers that are interested in metagenomics but have little experience, where the service is a standalone package in its own right .
For more information, check out the Metagenomics Service Description.
- Why do you require a gel image of my metagenomic DNA?
Metagenomic DNA extraction is no small feat. Often you are subjecting your sample to mechanical and chemical lysis to target as many different cell types as possible. However, an unintended side effect of this is degradation of the DNA during this process.
Highly degraded DNA will fail to sequence, and we know all too well the frustration of spending a long time in the lab only to get null results. So to make things as smooth as possible, we ask for you to check the integrity of your DNA before you send it to us so that both us and you are confident your DNA will sequence successfully.
Sometimes you can’t re-extract samples, and we’re happy to try and sequence it anyway, but in these cases we will not resequence any failed samples. We’ll ask you to agree to this before we proceed.
- What outputs do I get from your metagenomics services?
Customers using our metagenomics services will get access to:
- Raw data from sequencing and taxonomic assignment
- Taxonomic distribution tables
- Visualisations comparing sample diversity
All of your data will be delivered via our secure project portal.
To find out more, check out our Metagenomics Service Description.
- Can you do my metagenomic DNA extractions for me?
No, we currently only accept pure, intact DNA. We are working on offering metagenomic extractions, but the diversity and complexity of metagenomic sample types is vast.
Bioinformatics
- What bioinformatics analyses will you provide?
All of our services include a suite of bioinformatics analyses, meaning you can go straight from sample, to actionable results.
The types of analyses differ between our services, here’s a summary:
Whole Genome Sequencing Services
You’ll get an an assembly and annotation of your organisms, as well as a comprehensive suite of QC metrics, including taxonomic classifications of your reads and assemblies so you can be sure your sample is what you’re expecting.
Metagenomics Services
Our metagenomics services provide a granular taxonomic classification of your sample to several taxonomic ranks (Species, Genus, Phylum). Depending on how much metadata you provide to us, we will also generate principal component graphs to show how the taxonomic diversity and composition of samples is influenced by different variables.
- I just want the raw data. Can I opt out of the bioinformatics analysis?
No, all samples are put through the same bioinformatics pipeline, which forms part of our industry leading QC procedure, and is provided at no additional cost. You will of course still receive your raw data when it’s passed all of our checks.
- I need additional bioinformatics analysis. Can you help?
MicrobesNG does not offer a bespoke analysis service. We’re only a small team! But our bioinformaticians may be able to suggest relevant software or pipelines based on their extensive experience - get in touch and we’ll see what we can do.
- My Illumina data includes reads labeled U1 and U2, what are these files?
We perform quality trimming on your fastq data. As Illumina has forward and reverse reads, sometimes just one of the reads in either the forward or reverse orientation gets trimmed, leaving an 'unpaired' read in the corresponding file.
So these orphaned unpaired reads are split out into their own files. You’ll see four files for each sample, suffixed with 1, 2, U1 or U2, which indicate the forward read, reverse read, unpaired forward, and unpaired reverse respectively.
You’ll be primarily interested in the forward and reverse reads for your own analyses, but we still provide the unpaired reads in case it helps.
General
- Should I acknowledge MicrobesNG in my publication and if so how?
That’d be great! We love seeing what customers do with their data, and adding something like: "Genome sequencing was provided by MicrobesNG (https://microbesng.com)." in your methods will help us find and enjoy your research.
- What are your Terms and Conditions of Service?
Our Terms and Conditions of Service can be accessed here.
- What are your data processing terms?
The personal data we process is that which you provide to us. This is generally gathered from the completion of the online Quote Request Form, but may also be taken from other communications such as email, telephone and conferences.
The communications we send you will be tailored to any preferences you have expressed or derived from your use of our services to date. In some instances MicrobesNG may become a Data Processor. Full details can be found in our T&Cs.
MicrobesNG will process your personal data for the following purposes:
- To prepare formal quotes, progress orders and manage financial transactions.
- To communicate in relation to all samples and sample progress.
- To ensure ongoing communications regarding existing projects and enquiries.
- With your consent to provide you with communications on our services, events and promotions.
Information on how to exercise your right can be found through accessing our privacy information by following this link: Privacy Policy
- Who works at MicrobesNG, are you Hiring?
We're a small but dynamic team, and we're always reinvesting in the business to grow our strengths and capabilities.
You can see our current team here, and check for any job vacancies here.- My question isn't listed here?
Drop us an email info@microbesng.com, our friendly team will be happy to help!
Methods, Protocols & Documents
- Where can I access protocols and documentation?
You can access all of our relevant documents at microbesng.com/documents