Metagenomics DNA Requirements
The following checklist can be used as a quick reference to ensure your DNA meets our metagenomics submission criteria.
Samples that do not meet one or more of the criteria below can be refused, or fail to sequence entirely, so please ensure you read the entire document to avoid any issues or delays.
Volume: |
30 - 100 µL | |
Concentration ID Labels: |
1* - 30 ng/µL | |
Buffer: |
Does not contain EDTA | |
Integrity: |
Is not degraded | |
Tubes: |
Standard Microfuge Tube | |
Sample ID Label: |
PRINTED label applied to tube | |
Sample Info: |
Uploaded to Project Portal |
* ≥10 ng/µL if measured with spectrophotometer (e.g. NanoDrop)
If you are sending more than 20 DNA samples and you have indicated in your quote request that you will send them all together rather than in batches, we will ask you to send your samples in a 96-well plate using the instructions provided here.
Volume
We require a sample volume of 30 - 100 µL of DNA in an EDTA-free buffer.
Concentration
We require your DNA to be at a concentration of 1* - 30 ng/µL.
DNA samples must be quantified before sending them to us, and the concentration recorded in the sample sheet on your project portal. If you are sending multiple samples please normalise them to the same volume and concentration, this will speed up your turnaround time.
We strongly recommend the use of a fluorometric quantification method that is specific to DNA (e.g. Qubit, Quantus, Quant-iT, Picogreen).
If you do not have access to a fluorometric platform, you can use a spectrophotometric platform (e.g. NanoDrop) to quantify your DNA. However be aware that this modality often overestimates DNA concentration. Therefore, the minimum required concentration measured using a spectrophotometer is >10 ng/µL*.
Buffer
The best buffer to elute your DNA in is EB (10 mM Tris-HCl pH 8.5).
You can elute your DNA using ultrapure, nuclease-free water - in this case please ensure you store samples at -25°C to -15°C to prevent degradation.
We cannot accept DNA that is eluted in a buffer containing EDTA (e.g. TE buffer), as EDTA binds to bivalent cations (e.g. Mg2+) that are essential for enzymatic reactions in the library preparation process.
Integrity
You must check the integrity of all of your metagenomics samples.
This is important as overly fragmented samples will fail to sequence properly.
It is compulsory to submit proof of integrity of all your DNA samples before you send them by emailing info@microbesng.com. A member of our lab team will verify that your samples meet our submission requirements, and approve your samples for submission. Please include your project ID number in the email subject line.
If you do not submit proof of integrity, you will be given the option for us to accept your samples onto our sequencing pipeline, but we will not resequence any samples that fail due to sample integrity issues.
To check integrity, run your samples on an electrophoresis system like the Bioanalyzer HS DNA chip or Tapestation gDNA Screentape. Alternatively, you can perform gel electrophoresis with a 0.75% e-gel/agarose gel with a 1kb ladder.
An example of intact and degraded DNA is provided in Figure 1 below; non-degraded genomic DNA (Samples 1 + 2) will show a single band of High Molecular Weight (>40,000 bp). A smear between 100 - 40,000 bp indicates DNA degradation (Samples 3 + 4), which may jeopardise the success of our NGS library preparation.
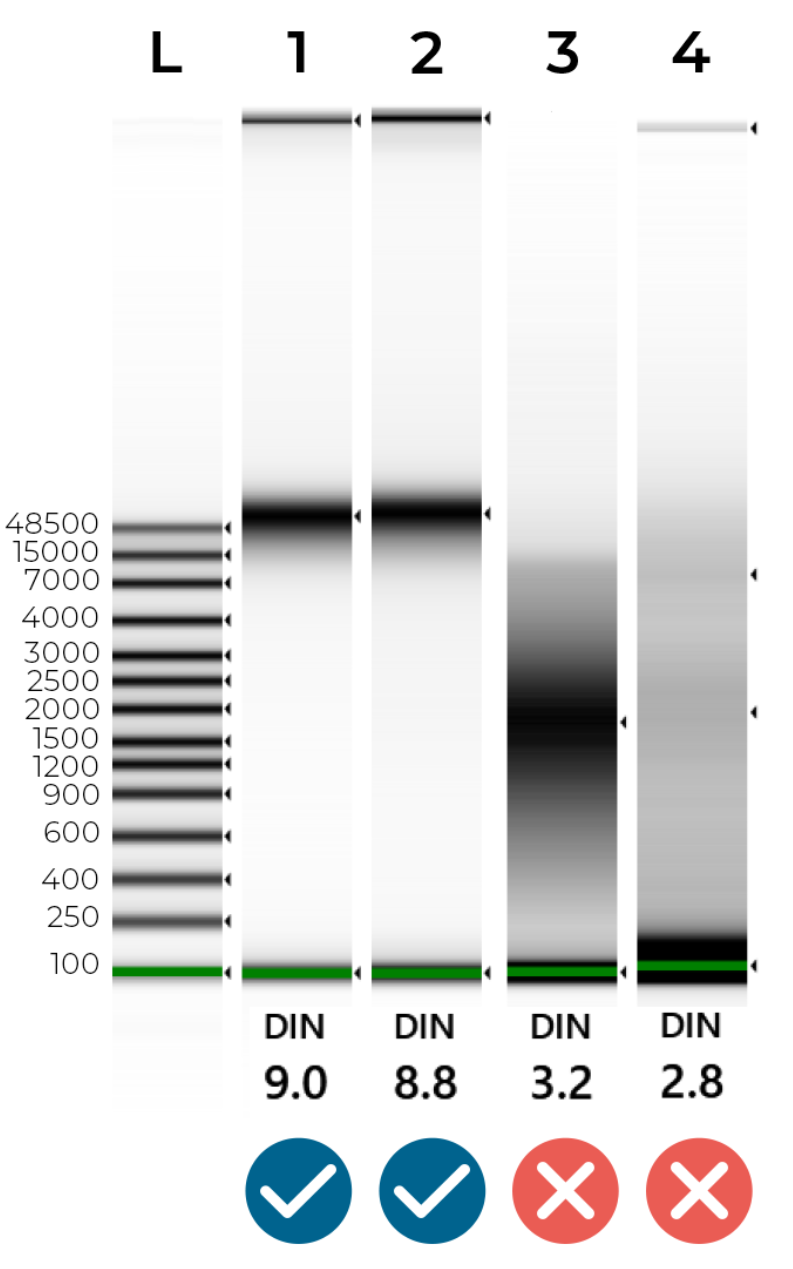 Figure 1 - Genomic Screentape results of intact and degraded DNA samples. The DIN (DNA Integrity Number) summarises DNA quality. Ideally, samples should have a DIN of 7.0-10.0. Samples are likely to fail sequencing if their DIN is lower than 6.0.
Figure 1 - Genomic Screentape results of intact and degraded DNA samples. The DIN (DNA Integrity Number) summarises DNA quality. Ideally, samples should have a DIN of 7.0-10.0. Samples are likely to fail sequencing if their DIN is lower than 6.0.
Tubes
DNA samples must be sent in standard microfuge tubes (i.e. tubes that fit in a regular benchtop centrifuge rotor), ideally with a screw cap lid, which offers the best security for your samples.
If you don’t have access to screw cap tubes, please put samples in 1.5 - 2 mL microfuge tubes wrapped in parafilm to minimize the likelihood of samples opening in transit.
We do not accept DNA sent in 0.5 mL or 0.2 mL tubes, or tubes that do not fit in a standard benchtop centrifuge, as they are incompatible with our workflow.
 Figure 2 - Acceptable DNA submission tubes.
Figure 2 - Acceptable DNA submission tubes.
Sample ID Label
You will be allocated unique sample ID’s that allow us to differentiate your sample and track it through our pipeline. This will be a 6 digit number (e.g. 239253)
Download the excel spreadsheet from your project portal and print your allocated MicrobesNG sample ID numbers and attach them to your sample tubes lengthwise, not around the tube (see Figure 3). If you don’t have a label printer, please use normal paper and a printer and attach the label to the tube with transparent tape (e.g. Sellotape). We can also send you preprinted labels if you prefer, but please note that this has an associated handling and shipping time.
Please do not handwrite your sample ID number on your tubes. Handwritten text can rub off, or be hard to interpret, so we do not accept tubes with handwritten labels.
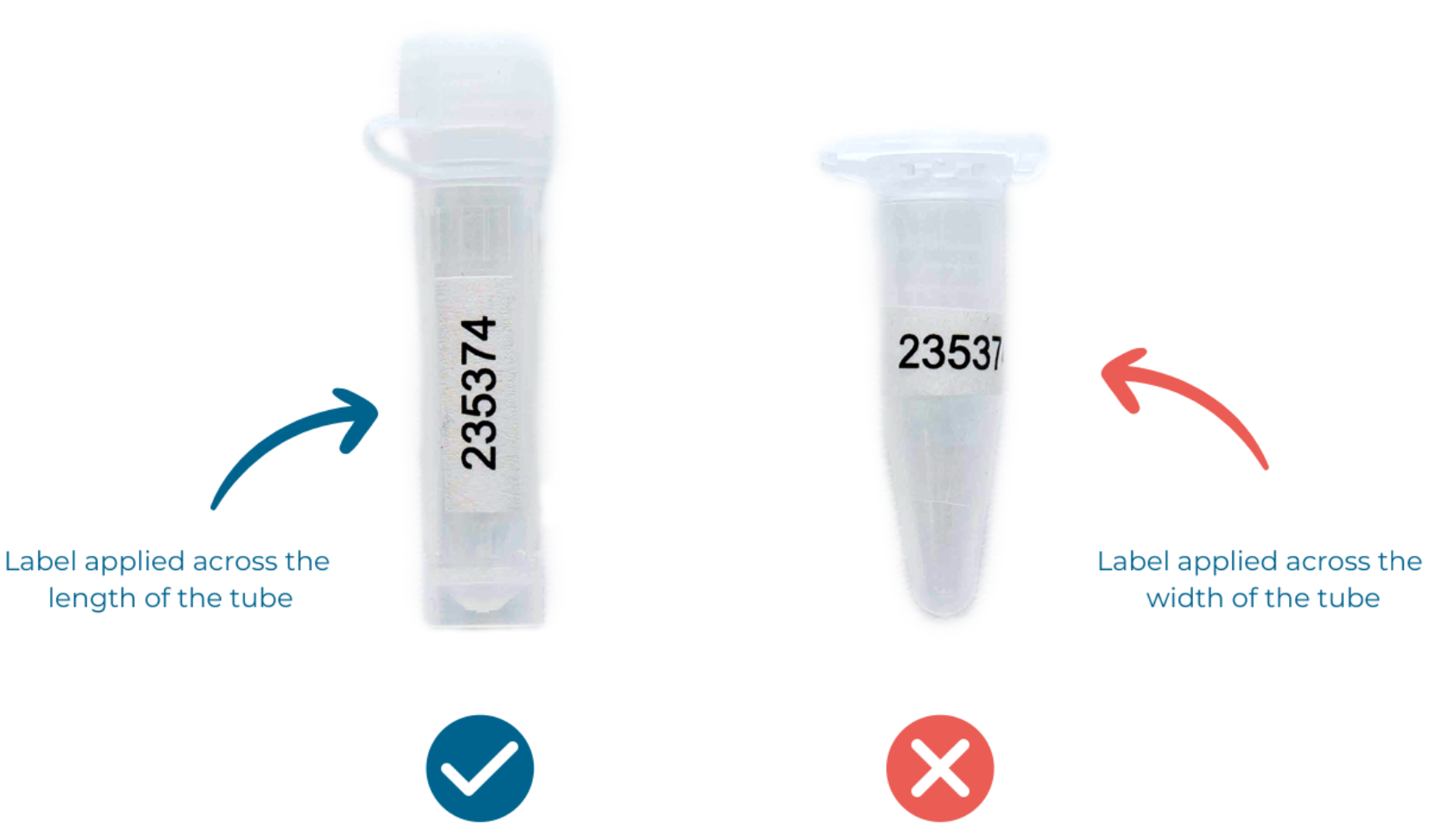 Figure 3 - How to correctly apply labels to your tubes. (Note: if you print your own labels only the sample ID number is required, not the barcode!)
Figure 3 - How to correctly apply labels to your tubes. (Note: if you print your own labels only the sample ID number is required, not the barcode!)
Sample Info
It’s really important that you fill in the metadata for your samples on the project portal before you post them. We use this information to optimise our library preparation, so we can not accept your samples without it.
If we do not have this information when your samples arrive in our lab, they will be placed on hold whilst we contact you, which could lead to a longer turnaround time.
Shipping
We recommend shipping your DNA at ~4˚C by including freezer packs or wet ice. For shipments that will arrive within 1-2 days, you can ship your DNA under ambient conditions.
To protect your samples from breakages and leaks, place them inside a rigid tube box, and place that box inside a rigid outer package (e.g. polystyrene or cardboard package).
For more information, see our Sending your samples guide.